De-intensification Effects on Methane-Cycling Microorganisms and the Methane Sink Function of Grasslands

Grassland soils are highly dynamic systems that can act as both sink and source for methane, the second most important greenhouse gas. Accordingly, both methane producing microorganisms, designated Methanogens, and methane consuming microorganisms, called Methanotrophs inhabit grassland soils. In the previous project (BE_CH4, 2017-2022), we showed that the role of grassland soils as a sink or source for methane can change within a year and that sink function is generally affected by land use intensity and nitrogen fertilization. In addition, we were able to link the gene expression of methane-cycling microorganisms to net surface fluxes of this potent greenhouse gas.
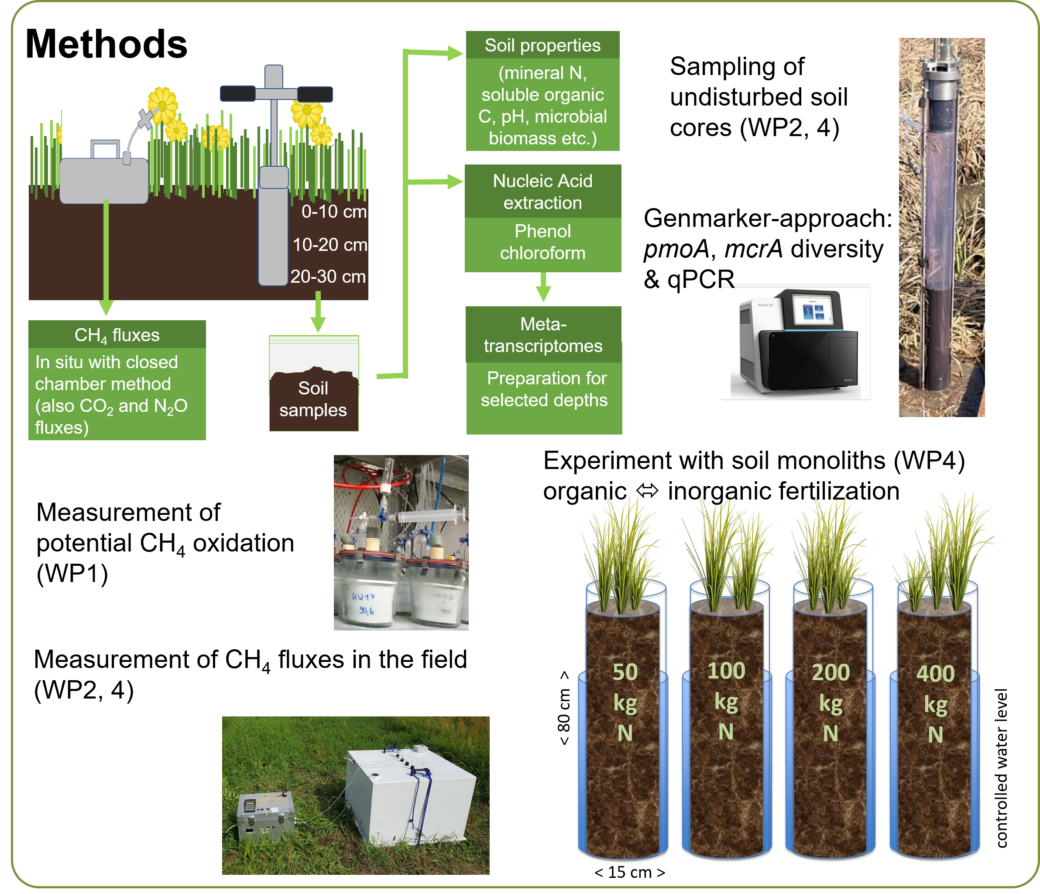
MetGrass aims to resolve effects of grassland management measures such as fertilization and mowing as well as long term de-intensification on methane-cycling microbiomes in top- and in subsoil. The project further aims to link these spatial functional biodiversity dynamics with methane net surface flux rates. MetGrass focuses on plots of the joint multi-site experiments REX and LUX and provides mechanistic experiments on soil monoliths on N fertilization levels.
We will quantify the abundance, diversity and activity of methane-cycling microorganisms with a gene marker approach by analyzing DNA/RNA encoding essential steps in methane cycling pathways. This microbiome data will be linked to methane net surface fluxes measured with the closed chamber method or potential methane oxidation rates determined through soil incubation. Information on soil physico-chemical properties and land use intensity will help us to unravel the effects of de-intensification of land-use on the methane sink function of grasslands.