BELongDead – Fungal Functional Diversity – Halle/Zittau/Grafenau Structural Diversity of Fungi and Bacteria in Deadwood and the Expression of Genes Relevant to Wood Degradation
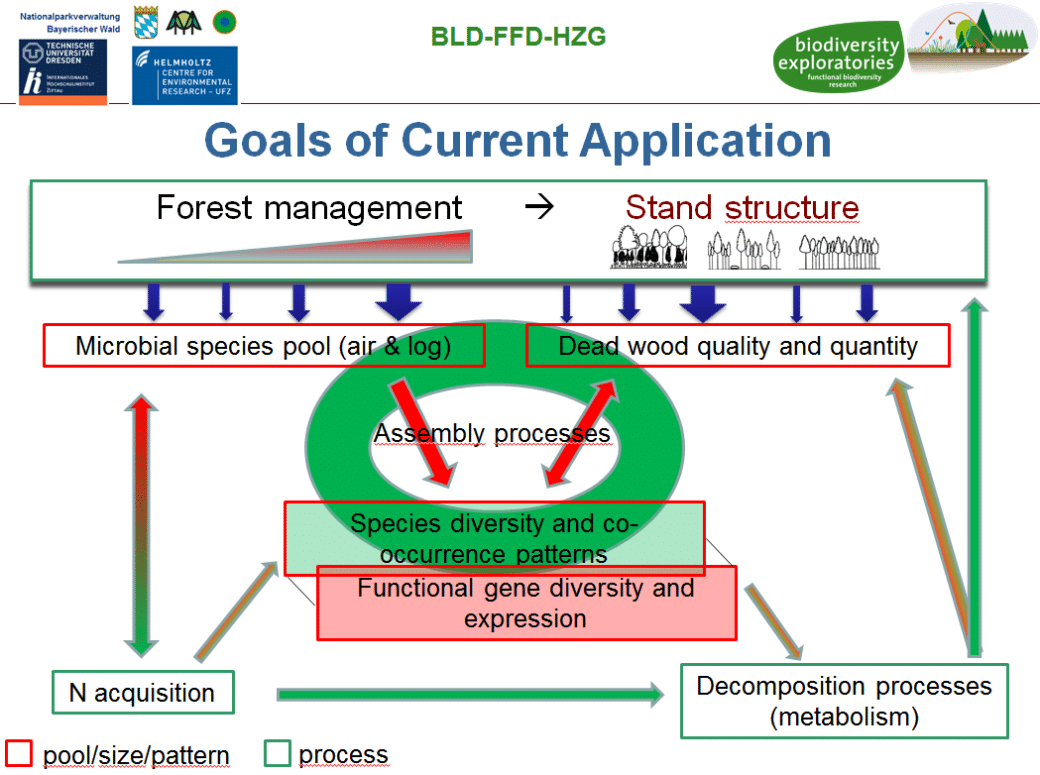
Compared to the predecessor projects FunWood I-II, which largely focused on the influence of forest management on microbial diversity in existing deadwood in decomposition under field conditions, our focus now shifts to an experimental platform. The BELongDead experiment was initiated in 2008 under the leadership of Prof. Dr. E.D. Schulze (MPI Biogeochemistry Jena) with the aim of investigating the influence of the surrounding habitat on deadwood and its decomposition processes. Another focus is on the long-term observation of the colonisation of deadwood logs by a wide variety of organisms.
It will be investigated to what extent I) the surrounding ecosystem influences deadwood decomposition, II) how deadwood colonisation occurs and III) how microorganisms control deadwood decomposition and thus influence ecosystem processes such as nutrient turnover. BELongDead allows us to study the influence of land use in the form of forest management on a standardised set of deadwood (12+1 different tree species of the same size and decomposition onset), evenly distributed in 3 replicates in the 3 exploratories and in 3×3 differently managed plots each.
The aim of our project is to combine state-of-the-art molecular biological methods with classical fruiting body mapping and spore collections to I) determine the type and quantity of wood decomposition and different forest management aspects, II) observe and study distribution and succession patterns of fungi over time, III) determine fungal activity at the transcriptome and enzyme levels and interweave these results with process data, IV) determine resulting changes in wood chemistry, V) determine the influence of N-fixing bacteria on fungal diversity, and vi) ultimately identify key species in these complex processes.
1. increased forest management intensity reduces the species pool of wood-inhabiting fungi at landscape and forest stand level.
2. intensive forest management is a habitat filter that favours certain species with certain life strategies (e.g. generalists).
3. forest management relaxes competitive interactions between wood-dwelling fungi, leading to higher wood decomposition rates.
4. wood decomposition processes are highly predictable from fruiting body dating combined with molecular data on phylogenetic and functional diversity.
5. there is consistent microbial N-fixation in deadwood, and certain bacteria occur non-randomly with certain fungi.
6. among the typical peroxidases expressed by fungi (Class II peroxidases, hem-thiolate peroxidases), we expect manganese peroxidases to be the most diverse and abundant of all lignin-modifying enzymes.
7. peroxidase transcriptome diversity increases with greater diversity of white rot fungi.
To answer our questions, we use state-of-the-art molecular biological techniques on DNA and RNA level, including so-called “Next Generation Sequencing” techniques (NGS). Furthermore, we want to prove the actual N-fixation in the wood and allocate the N to the corresponding participants. For this purpose, we are planning a SIP experiment (“stable isotope probing”), together with NGS metagenomics, real-time PCR and bioanalytics (acetylene reduction tests, mass spectrography).
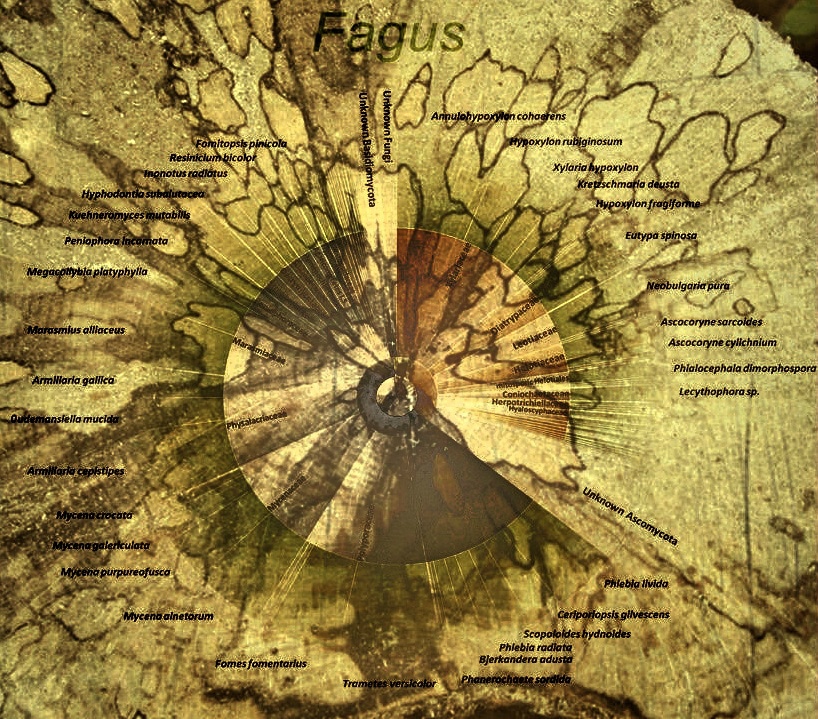
In addition to intensive fruiting body mapping, spore collectors will also be installed to identify airflow-based distribution patterns and to compare which fungi are potentially present (species pool) and which are already established in the deadwood.