BELongDead – Multitrophic functional diversity in deadwood (BLD-MFD-HZG III)
Deadwood is an important source of organic carbon and nutrients in forest ecosystems, which is continuously degraded by macro- and microorganisms, which hereby structure the ecosystems and impact multiple ecosystem functions. Though many contributions on species diversity for single taxonomic groups in relation to various deadwood substrates have been made through the last years, we still lack a comprehensive understanding of how wood-inhabiting species are organised, assemble over time, and specifically contribute to wood decomposition processes.
Our project is part of the BELongDead (Biodiversity Exploratories Long-term Deadwood) experiment that elucidates the decomposition of deadwood logs of thirteen deciduous and coniferous temperate tree species, standardised by the same starting time point of decomposition. We intend to quantify all relevant underlying molecular and biochemical mechanisms and processes of microbial-mediated wood decomposition in relation to forest management practices and geographical scale.
Particularly, we analyse:
- How species colonise a deadwood object
- How communities from different taxa interact during succession
- How the importance of host characteristics and environment change during succession
- Whether the relationship between diversity and decomposition can be better understood by considering multiple taxa
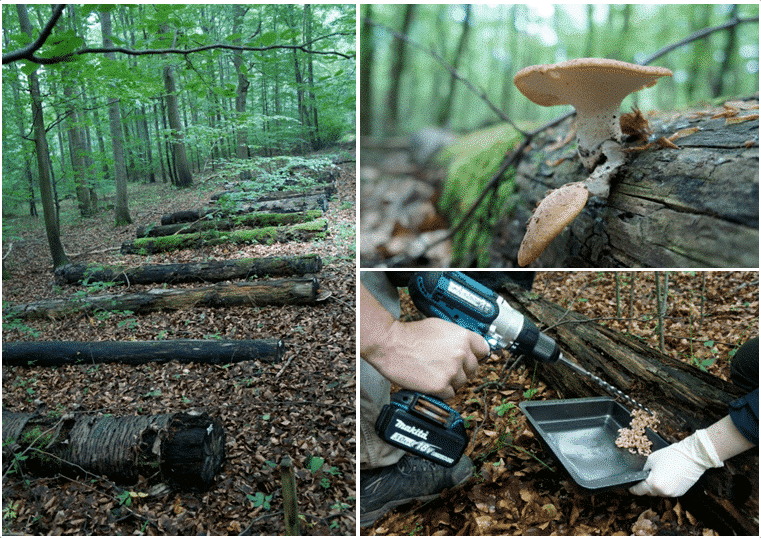
We use classical sampling methods and state-of-the-art molecular tools to capture as many aspects of diversity und community structure (Fig. 1). This information will be linked to wood chemistry, activity patterns of decomposing enzymes and wood decomposition in general. We collect the following data on two experiments:
- DNA-based diversity of fungi, bacteria, archaea and nematodes
- Diversity of fungal fruit bodies, lichens and mosses (Fig. 2)
- Wood characteristics, such as pH value, C/N ratio, lignin content
- Activities of wood-degrading enzymes (e.g., peroxidases, laccases)
These data are collected in two different experiments:
a) BELongDead
After sampling in 2012, 2015 and 2017, our data series will now be extended by a fourth time point in 2020 to enable assessing the processes of wood decomposition from the initial to the sometimes far advanced decomposition phase.
b) BESterile
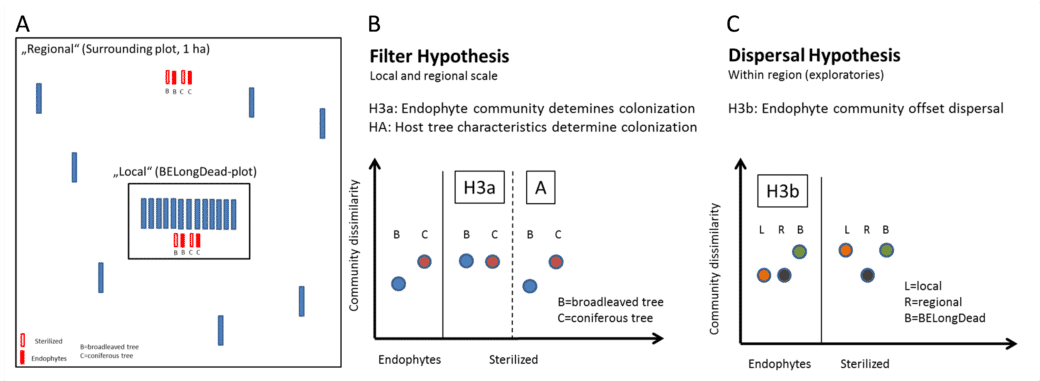
- In a new additional experiment, gamma-sterilised and untreated deadwood logs of the two main tree species of Central European forests Fagus sylvatica and Picea abies are laid out next to the existing BELongDead logs and at a precisely defined distance (Fig. 3A). In this way, we want to answer two fundamental research questions:Does a comparison between the unsterilised stems (characterised by the endophyte community) and the sterilised controls show a difference in colonising diversity and, inferentially, in wood decay rate? Are the differences between different tree species mainly caused by the endophyte communities (Fig. 3B, hypothesis H3a) or alternatively by host tree characteristics (e.g., physicochemical properties of the wood, Fig. 3B, hypothesis H3A)?
- We expect colonisation to depend on the donor pool and thus differences in colonisation patterns between stems directly at BELongDead (stronger on the local species pool) and those further away (stronger on the regional species pool). If the endophytic community determines colonisation, we will observe a more similar species community between all strains (Fig. 3C, hypothesis H3b).