BELongDead – Multitrophic functional diversity in deadwood (BLD-MultiFuncDiv)

Deadwood is an important source of organic carbon and nutrients in forest ecosystems and is continuously degraded by macro- and microorganisms. The decomposition of deadwood contributes to ecosystem structuring and impacts multiple ecosystem functions. Though many studies on species diversity for single taxonomic groups in relation to various deadwood substrates have been made through the last years, we are still lacking comprehensive understanding how wood-inhabiting species are organized and assembled over time and how they specifically contribute to wood decomposition processes.
Two real-world experimental set-ups within the Biodiversity Exploratories provide a unique opportunity to fill important knowledge gaps related to the organismic-mediated decomposition process:
(I) The BELongDead (Biodiversity Exploratories Long-term Deadwood) experiment investigates the decomposition of 13 temperate deciduous and coniferous tree species and the environmental drivers affecting this ecosystem process. This allows us to better understand the mechanisms of species diversity and community assembly during (partly) complete decomposition in relation to forest management intensity and forest structure.
Here, we analyse:
- a) how species colonize a deadwood object,
- b) how communities from different taxa interact during succession,
- c) how the importance of host characteristics and environment change during succession and
- d) whether the relationship between diversity and decomposition can be better understood by considering multiple taxa.
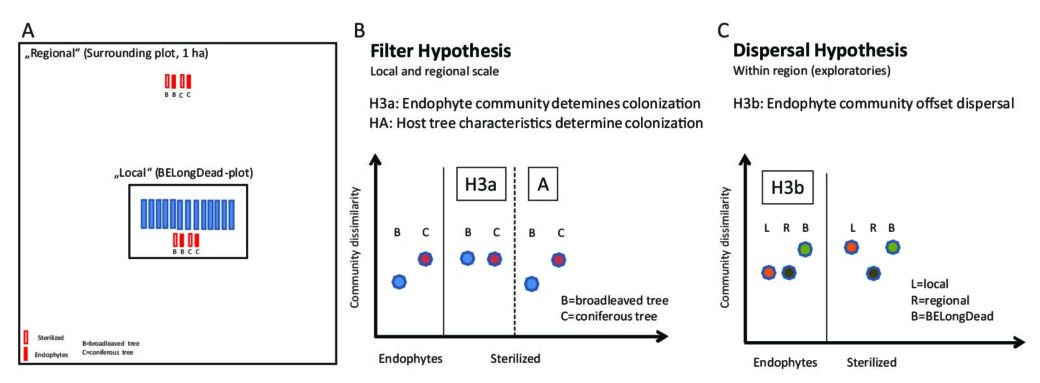
(II) Based on BESterile (Biodiversity Exploratories deadwood Sterilization experiment), we focus on the mechanisms of species colonization and related processes under different forest management intensities in the early successional phase. For this purpose, the logs of two main tree species within Central European forests (Fagus sylvatica and Picea abies) have been gamma sterilized or non-sterilized (harboring natural endophytes) and then placed next to the existing BELongDead logs or in a well-defined distance (see Fig. 1A).
Here, we analyse:
- a) A comparison between the sets of non-sterilized logs (characterized by the endophyte community) and sterilized (controls) allow testing and answering the question whether observed and anticipated differences of the fungal diversity between different tree species is caused mainly by the endophyte communities (Fig. 1B, Hypothesis H3a) or alternatively by host tree characteristics (e.g. wood physico-chemical properties, Fig. 1B, Hypothesis H3A).
- b) We expect that the probability of colonization depend on the donor pool. If this holds true, we would observe significant differences in colonization patterns between the logs that are exposed directly on the BELongDead plots (more exposed to the local species pool) compared to logs that are exposed in a certain distance (‘away’, more exposed to the regional species pool. If the endophytic community offsets the effect of the donor pool, we would observe a similar community between all logs of the same tree species (Fig. 1C, Hypothesis H3b).
I) BELongDead
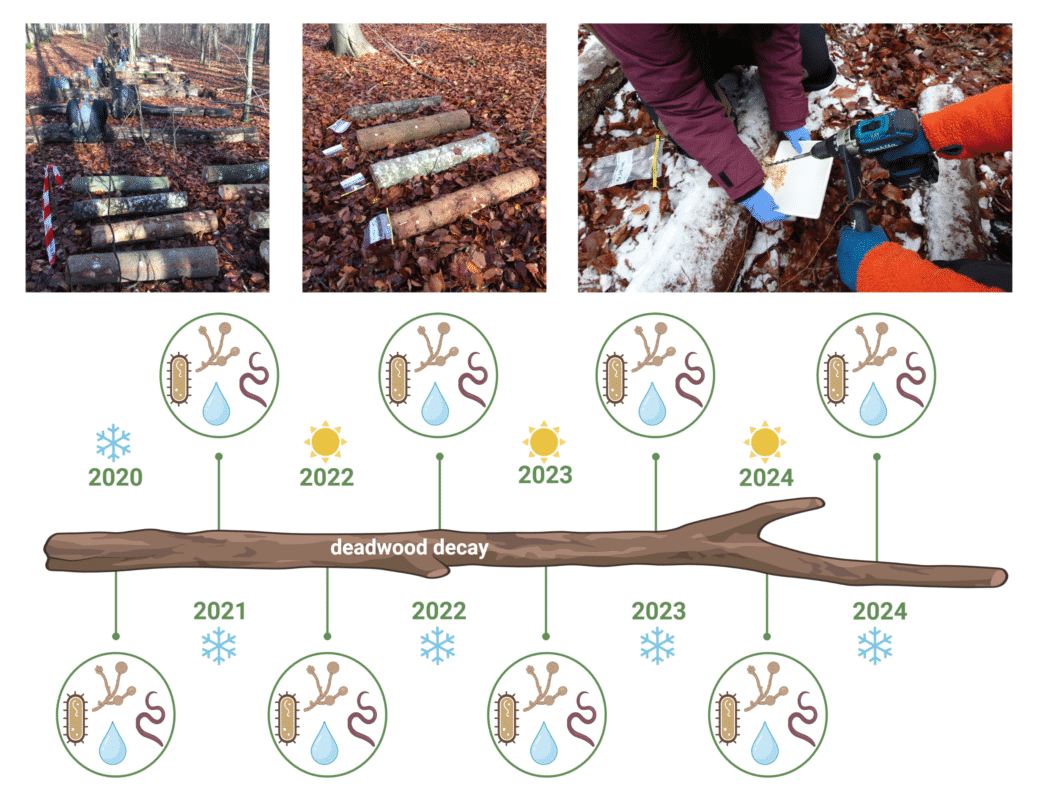
After wood sampling in 2012, 2015, 2017 and 2020, our data set is currently extended by a fifth time point in 2023 in order to evaluate the processes of wood decomposition from the initial to the very advanced decomposition phase (Fig. 2). For each time point we have investigated the deadwood-inhabiting fungal and bacterial community by amplicon sequencing using standardized protocols. These data are completed by different wood characteristics, degrading enzyme activities, and the diversity of fungal fruiting bodies, archaea & nematodes for several sampling campaigns (Fig. 3).
I) BESterile
Since autumn 2020 (T0) wood samples have been collected twice a year according to the standardized BELongDead protocol. These samples are used to investigate the bacterial, fungal & nematode diversity via amplicon sequencing and basic wood characteristics such as pH and water content (Fig.4).