Drivers of recruitment and diversity in symbioses: Assessing changes in fungal, algal, and bacterial communities of the lichen holobiont along land-use gradients
Forest environments are home to diverse communities of microorganisms like algae, fungi and bacteria in a variety of environmental pools. Representatives of these microorganismal groups can be found on virtually any substrate, including soil and tree bark. Lichens are one of the oldest known examples of symbiotic organisms and are traditionally viewed as mutualistic symbioses between a fungus and a green alga or cyanobacterium. However, the discovery of numerous additional microorganisms that potentially act as obligatory participants in the symbiosis, has challenged this assumption.
Nowadays lichen individuals are regarded to be complex ecological units (holobionts), harbouring diverse communities of fungi, algae as well as bacteria. These ‘miniature ecosystems’ can be used to shed light on community assembly processes, species interactions, or concerted responses to the environment at the micro-level. So far, we know little about interactions and relationships between microbial communities from different, but physically proximate environmental substrates, like lichen holobionts, bark biofilms and soil. This is mainly due to the absence of extensive microbial diversity data from these niches. An exhaustive understanding of biodiversity is the first essential step toward testing how forest features and land-use intensity affect community assembly processes of these interacting microorganismal groups.
We want to understand how forest features and forest management practices affect diversity and community structure of microorganisms (fungi, green algae, bacteria) in forest environments. We will thus assess these communities on bark, in bark-associated lichens, and in soil. Our study will provide insights into the frequency and stability of biotic interactions among different microorganismal groups along environmental gradients, recruitment processes of microorganisms into the lichen holobiont, and above ground – below ground community interactions.
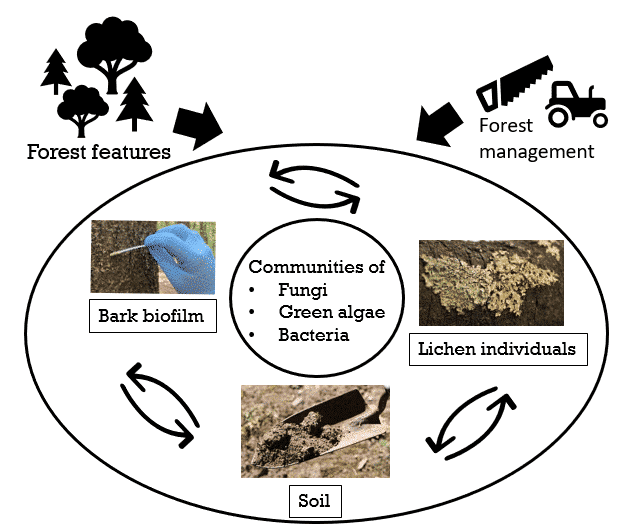
- Obtain samples of bark biofilm and soil (cooperation with core projects soil and microorganisms) from all 150 EP forest plots. Obtain epiphytic lichen thalli from selected forest plots.
- DNA extraction and PCR of fungi (ITS), green algae (ITS) and bacteria (16S).
- High-Throughput metabarcoding (Illumina MiSeq) to assess microbial diversity.
- Examine overlap in species composition, e.g., between bark biofilm, soil and lichen holobiont.
- Model responses of diversity and biotic interactions to forest features along a land-use gradient.